One-click exploration of AiiDA archives via RenkuLab#
Did you know that when you conduct research using AiiDA, you can host the generated data for free on the Materials Cloud Archive (MCA)? By doing so, you make your data available to the research community at large, offload the responsibility of long-term data storage, and allow users to explore it with MCA’s built-in provenance explorer.
Tip
Try it now! Want to see this in action? Head to the Materials Cloud Archive, find any record on MCA (e.g., the “Verification of DFT precision via reproducible workflows” one), and click the Renku logo to launch a fully configured environment!
The Problem: No data access without local setup#
Sounds great, now, where’s the catch? While uploading the data to MCA makes it, in principle, available to the world, exported AiiDA archives are not directly human-readable. This is because they are not just classical directory (and file) trees, but compressed files with a specific format that was designed for performance[1]. Therefore, exploring the data is not straightforward, and typically required users to install AiiDA locally (including complex system dependencies like PostgreSQL and RabbitMQ[2]), download the archive, and then use AiiDA’s API and/or CLI to explore it.
This barrier meant that the data hosted on MCA was effectively only available to users already familiar with AiiDA or willing to invest the necessary time in learning it. And, still, even experienced users had to manually download the archives and configure their local environments before exploring the data—time that could have been spent on actual research instead.
The Solution: Integration with RenkuLab#
Hence, the AiiDA development team decided to partner with RenkuLab, a platform developed at the Swiss Data Science Center (SDSC) for reproducible and collaborative data science, to address these challenges. Both tools share a Swiss origin and a strong commitment to reproducibility in science, making them natural partners. The first version of an integration between the two platform was established as early as 2019[3].
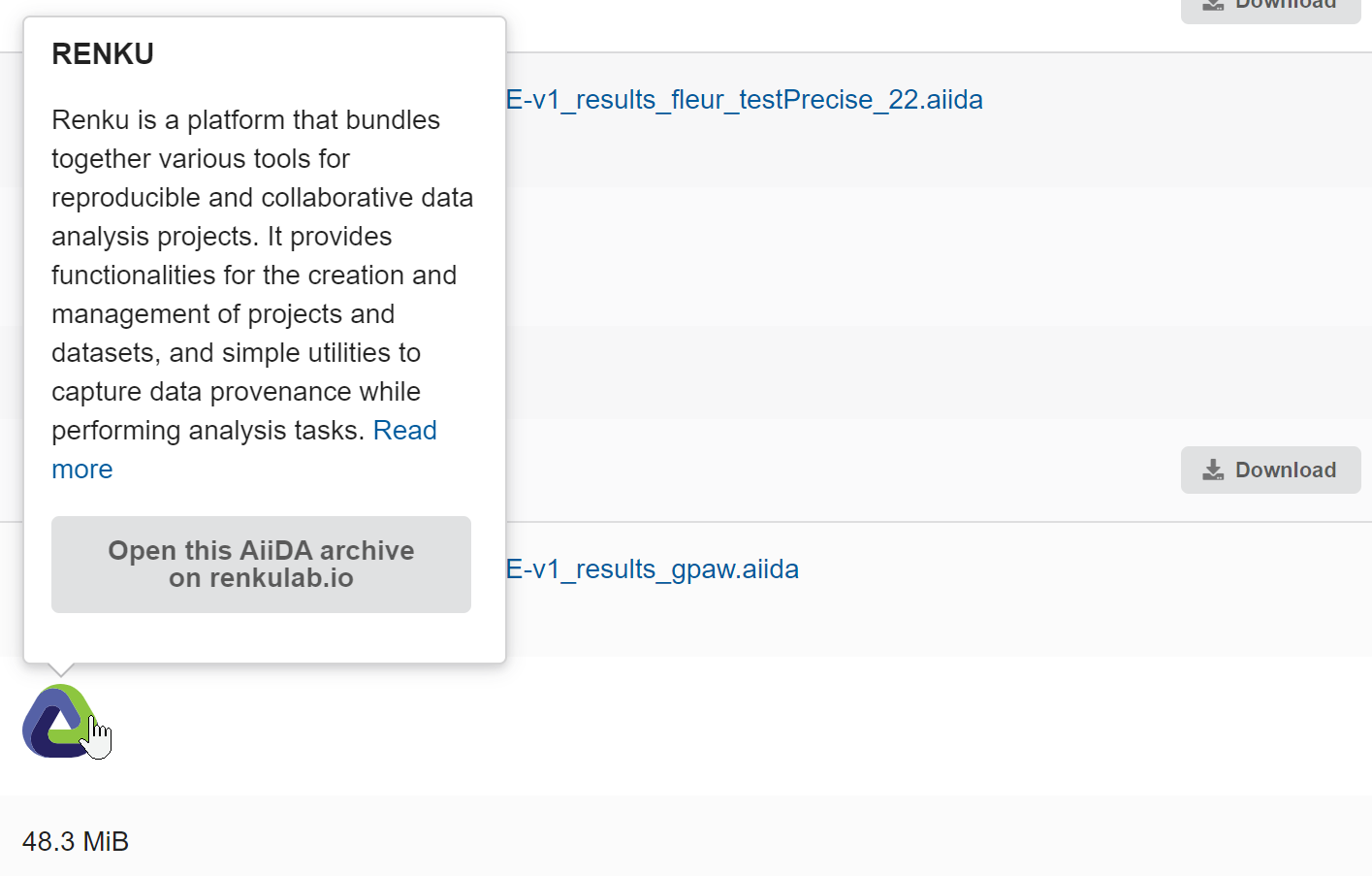
It allows researchers to click the Renku logo next to any AiiDA archive on MCA and be automatically launched into a fully configured RenkuLab project with AiiDA pre-installed and the data ready for exploration, thus eliminating the need for any local installation or configuration:
See also
Before diving into the technical details of the integration, note that the Renku team has also written about this collaboration from their perspective in a blog post on their website.
Initial Implementation (Renku 1.0)#
The first implementation was built as a contributed Renku template and was based on a Renku Docker image, with the AiiDA installation baked into the image creation process.
Jinja2 placeholders were used to conditionally populate metadata in a startup Jupyter notebook, and other configuration files, based on the URL of the MCA record from which the Renku project was launched[4].
A post-init.sh script, for example, set up the AiiDA profile using the appropriate storage backend (sqlite_zip) to directly mount the provided archive in read-only mode.
On the MCA side, the integration was achieved by building a Renku project launch link with all the necessary parameters to automatically configure the template based on the selected archive. This link is accessible via the Renku logo displayed next to each AiiDA archive on the MCA record pages (see image above). The initial implementation still required user interaction with Renku’s graphical user interface (GUI) during the project creation process, though all data was pre-filled based using the data contained in the launch link.
Current Implementation (Renku 2.0)#
With the upgrade to Renku 2.0, the current implementation retained its core technical foundations from the first version, but introduces several key improvements: For one, the GUI-based project creation step required in Renku 1.0 is bypassed through the use of a pre-created project, enabling genuine one-click AiiDA archive access from MCA.
In addition, archive download and AiiDA profile creation has been deferred to manual execution of notebook cells by the user, rather than being done at session startup. This drastically reduces startup time, as downloads of multi-gb archives could previously take minutes during session startup.
Further, in Renku 2.0, the archive URL that points to the MCA record from which the session was launched is available as an environment variable and is used to fetch relevant metadata via the InvenioRDM API[5]. This allowed for a more customized startup notebook with the set of displayed cells adapted based on the MCA entry.
The startup notebook now also contains additional code snippets showcasing AiiDA’s API for archive exploration, while internal backend code (e.g., archive download and AiiDA profile setup) is hidden in auto-folded cells.
Finally, the Docker image now includes the latest released aiida-core version, 2.7.1, with all its recent improvements. Here, one notable feature is the capability to dump all relevant data of an AiiDA process, group, or profile to disk in a human-readable directory and file tree[6]. This data can then be explored via the Jupyter file browser, command line utilities, or downloaded to the local machine, greatly simplifying data access also for non-AiiDA users. Instructions on how to use the feature are contained in the startup notebook, as well.
A visual journey of the integration
Below, we show the full journey from an archive entry on MCA to the working RenkuLab project, going through the startup notebook, the code examples, and finally exporting the AiiDA data to a directory tree. Please note that some parts of the original recording were cut out to shorten the video (e.g., session startup on Renku typically takes approx. 1 minute):

Future Directions#
In the development team, we are already exploring several enhancements using Renku 2.0’s new capabilities.
Data Connectors
Renku 2.0 provides “data connectors” that can mount data from supported providers directly in a RenkuLab project’s filesystem. Since MCA now makes use of InvenioRDM in the backend, which is supported by Renku 2.0, creating an MCA data connector could allow mounting AiiDA archives without needing to download them at all. This would further reduce the startup time and would also circumvent storage limitations typically applied by Renku (every project comes, by default, with 10GB of disk space).
One technical challenge currently still prevents the use of data connectors: many MCA archives were created with older AiiDA versions and require data migrations to work with current versions of AiiDA. Since mounted filesystems via data connectors are read-only, AiiDA’s current approach of in-place migration doesn’t work. Potential solutions include periodic, automatic migrations of MCA-hosted archives to the latest version of AiiDA, or creation of the migrated file on the RenkuLab project’s actual filesystem (though this sacrifices one of the main advantages of data connectors—instant access without the need for downloading the whole file).
Expanding the AiiDAlab App Ecosystem
RenkuLab 2.0’s support for arbitrary Docker images opens possibilities for integrating other AiiDA applications.
Work has already been carried out to make the AiiDAlab Quantum ESPRESSO (aiidalab-qe) app compatible with Renku[7], potentially providing the same GUI and user experience as the standalone app.
With this, we also plan to improve the aiida-archive-inspect aiidalab app, with which information about all AiiDA archives available on an MCA record page could be shown, and the selection could be done interactively via the GUI.
Conclusion#
The integration between AiiDA and Renku demonstrates how research infrastructure can evolve from serving primarily expert users to welcoming the broader scientific community. By removing barriers between data discovery and analysis, the integration enables more researchers to engage with the computational datasets hosted on MCA, ultimately accelerating scientific discovery in materials science and beyond.
Finally, we would like to mention that RenkuLab has been very stable throughout the various stages of the integration, and the Renku team has consistently been responsive and helpful with technical concerns. While the AiiDA-RenkuLab integration uses only a small part of Renku’s technical infrastructure, the platform’s ability to connect external code and data sources with ready-to-use computational environments makes it a great tool we recommend to every computational scientist.
For researchers interested in exploring AiiDA archives on the Materials Cloud Archive on Renku, simply look for the Renku logo next to any archive and click it to begin your exploration. Have fun! 🚀